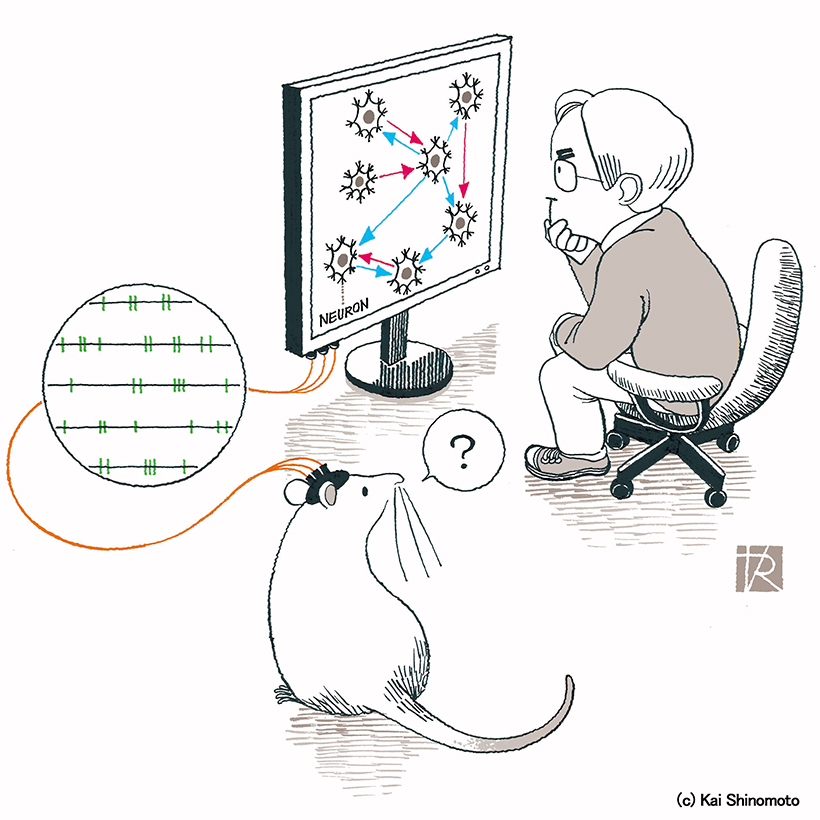
Reconstructing neuronal circuitry from spike trains.
This application program estimates inter-neuronal connections from parallel spike trains, based on the generalized linear model applied to cross correlation (GLMCC) developed in Nature Communications (2019) 10:4468, or its revised version. The references are in the below. You may also try a comprehensive application program that also contains an application CoNNECT.
Prepare your data in a {.txt} format. The data consists of a set of neuronal spike trains separated by semicolons {;} as
{spike train of the 1st neuron};
{spike train of the 2nd neuron};
...
{spike train of the Nth neuron};
Each {spike train} is given as a series of spike times separated by a newline, a comma, or a space. A sample in which spike times are separated by a newline (and spike trains are separated by a semicolon) is shown below
1692.529986 2372.809986 2682.789986 ... ; 1396.319986 1405.629986 1713.209986 ... ;
In our default setting, spike times should be represented in a unit of [msec], but you can change the setting into [sec] or [µsec].
other parameters
other parameters
Setting
(1) tau=1ms: time scale of interaction kernel was chosen 1ms.
(2) min_s = 1: an interval of ± 1 ms in the cross-correlogram was excluded, because near-synchronous spikes were not detected in this experiment due to the shadowing effect (p6 in the paper).
(3) ds = 1 2 3 4: synaptic delay at each connection was selected automatically.
unit of time
cross-correlogram: